AI Research Review 25.02.21 – AI for Science
Evo2 genomic foundation AI model, AI co-scientist accelerates science, · AI for modelling infectious disease epidemics.

Introduction
The greatest potential for AI to change the world is its ability to accelerate science and technology progress. This week’s AI Research Review covers three AI research papers that apply AI to biology, science research, and disease epidemics:
Evo 2: Genome modeling and design across all domains of life
Accelerating scientific breakthroughs with an AI co-scientist
AI for modelling infectious disease epidemics
Evo 2: Genome modeling and design across all domains of life
Its deep understanding of biological code means that Evo 2 can identify patterns in gene sequences across disparate organisms that experimental researchers would need years to uncover. The model can accurately identify disease-causing mutations in human genes and is capable of designing new genomes that are as long as the genomes of simple bacteria. – From Genome modeling and design across all domains of life with Evo 2
Arc Institute researchers have developed a foundation AI model for biology called Evo 2 that is trained on the DNA of over 100,000 species across the entire tree of life. They published a pre-print “Genome modeling and design across all domains of life with Evo 2” and also released Evo 2 as an open-source model on Nvidia’s BioNeMo.
Evo 2 is an improved version of Evo and a suite of two models, 7B and 40B parameters, that models DNA sequences at single-nucleotide resolution at up to 1 million base pair context length. Evo 2 is based on the StripedHyena 2 convolutional hybrid architecture, that combined transformers and StateSpaceModels (SSMs).
Evo 2 was trained on the OpenGenome2 dataset, which contains 8.8 trillion tokens of genomic data from all domains of life. Evo 2 7B was trained on 2.4 trillion tokens and Evo2 40B trained on 9.3 trillion tokens. Both were trained in two phases, first at 8,192 token context length to learn functional genetics, then they extended Evo 2’s context length to 1 million tokens to capture organism-scale genomes.

This genomic training and 1 million base pair context length enables Evo2 to perform genome modeling and design across different scales and across all domains of life:
Evo 2 learns from DNA sequence alone to accurately predict the functional impacts of genetic variation—from noncoding pathogenic mutations to clinically significant BRCA1 variants—without task-specific finetuning. Applying mechanistic interpretability analyses, we reveal that Evo 2 autonomously learns a breadth of biological features, including exon–intron boundaries, transcription factor binding sites, protein structural elements, and prophage genomic regions. Beyond its predictive capabilities, Evo 2 generates mitochondrial, prokaryotic, and eukaryotic sequences at genome scale with greater naturalness and coherence than previous methods.

Evo2 is a huge boost to biological science and the study of genetics. Evo2 is fully open source, with the Arc Institute sharing model parameters, training code, inference code, and OpenGenome2 training dataset to the public domain. Evo 2 is available to use via ArcInstitute’s Evo Designer interface and Nvidia’s BioNeMo service.
Accelerating scientific breakthroughs with an AI co-scientist
Google researchers have introduced AI co-scientist in a blog post and in a research paper titled “Towards an AI co-scientist.” AI-coscientist is a multi-agent system built on Gemini 2.0, designed to augment the scientific discovery process by collaborating with scientists to uncover new knowledge and formulate novel research hypotheses and proposals.
The AI co-scientist’s design is inspired by the scientific method and incorporates a “generate, debate, and evolve” approach to hypothesis generation, accelerated by scaling test-time compute. The system bears similarity to Sakana’s AI Scientist as well as the Deep Research tools developed by Google and OpenAI, but goes much further to fully support a rigorous research process:
Given a research goal in natural language, the co-scientist generates novel research hypotheses and proposals. The system employs specialized agents -Generation, Reflection, Ranking, Evolution, Proximity (which evaluates relatedness), Meta-review (which provides high level analysis) - to continuously generate, debate, and evolve research hypotheses within a tournament framework. Feedback from the tournament enables iterative improvement, creating a self-improving loop towards novel and high-quality outputs.
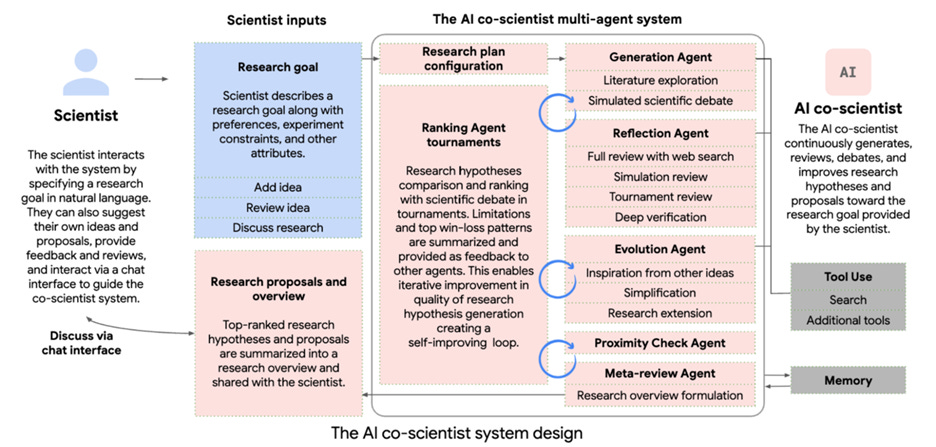
The system was validated in three biomedical areas - drug repurposing, novel target discovery, and explaining mechanisms of bacterial evolution and antimicrobial resistance:
In drug repurposing, the AI co-scientist proposed drug candidates for acute myeloid leukemia that show promising validation findings, including tumor inhibition in vitro at clinically applicable concentrations.
In novel target discovery, the AI co-scientist proposed new epigenetic targets for liver fibrosis, validated by anti-fibrotic activity and liver cell regeneration in human hepatic organoids.
In explaining mechanisms of bacterial evolution and anti-microbial resistance, the AI co-scientist recapitulates unpublished experimental results via a parallel in silico discovery of a novel gene transfer mechanism. The result was published as a paper.
The AI co-scientist was able to creatively assist in new scientific discoveries that became published results. As such, it presents a key advance in AI-assisted augmentation of scientists and acceleration of scientific discovery. The authors conclude that the AI co-scientist has the potential to meaningfully accelerate scientists' endeavors to resolve grand challenges in human health, medicine, and science.
AI for modelling infectious disease epidemics
A survey published in Nature called Artificial intelligence for modelling infectious disease epidemics examines how AI can be used to model infectious disease epidemics. This research demonstrates how AI can improve global preparedness for future pandemics, by improving human decision making in infectious disease epidemiology.
This review considers the use of AI in population health, applying AI systems that combine machine learning, computational statistics, information retrieval and data science to infectious disease modelling:
We first outline how recent advances in AI can accelerate breakthroughs in answering key epidemiological questions and we discuss specific AI methods that can be applied to routinely collected infectious disease surveillance data.
The study finds that recent advanced AI methodologies are performing increasingly well, even with limited data, currently a major bottleneck. Better performance on noisy and limited data is opening new areas for AI tools to improve health across both high-income and low-income countries.
The authors note that AI’s potential for pandemic disease mitigation is great, but that it depends upon worldwide collaboration and comprehensive surveillance data inputs.
In the next five years, AI has the potential to transform pandemic preparedness.
It will help us better anticipate where outbreaks will start and predict their trajectory, using terabytes of routinely collected climatic and socio-economic data. It might also help predict the impact of disease outbreaks on individual patients by studying the interactions between the immune system and emerging pathogens.
Taken together and if integrated into countries’ pandemic response systems, these advances will have the potential to save lives and ensure the world is better prepared for future pandemics. - Professor Moritz Kraemer, lead author of Artificial intelligence for modelling infectious disease epidemics



