AI Research Roundup 24.05.10
Dr Eureka, AlphaFold 3, xLSTM, KAN, Visualization-of-Thought, Agent Hospital
Introduction
Our AI research highlights for this week:
Dr Eureka: Language Model Guided Sim-To-Real Transfer Learning
AlphaFold 3: Predicting structure and interactions of life’s molecules
xLSTM: Extended Long Short-Term Memory
KAN: Kolmogorov–Arnold Networks
VoT: Visualization of Thought
Agent Hospital: Simulating a Hospital with Medical Agents
Dr Eureka: Sim-To-Real Transfer Learning with LLMs
Dr Jim Fan and colleagues at NVidia have been pioneering ways to train robot using simulated environments. Last year, they gave us Eureka, using LLMs to automatically generate reward algorithms for training robots. Now Dr Jim Fan shared Dr Eureka, from U Penn, that goes even further, with the paper “DrEureka: Language Model Guided Sim-To-Real Transfer.” What is DrEureka?
DrEureka [is] an LLM agent that writes code to train robot skills in simulation, and writes more code to bridge the difficult simulation-reality gap. It fully automates the pipeline from new skill learning to real-world deployment.
The basic idea is using LLMs to automatically define and run simulation environments to train robots in simulation, then transferring that learning into the real world. This make for efficient and scalable robot training.
Dr Fan notes that a dog-like robot balancing on a Yoga ball (see Figure 1) is hard because it is not possible to accurately simulate the bouncy ball surface. To train such a skill in simulation, domain randomization is used to define possible parameters for simulation. AI models like GPT-4, which “have tons of built-in physical intuition for friction, damping, stiffness, gravity, etc.” is used to define ranges of parameters.
Our LLM-guided sim-to-real approach requires only the physics simulation for the target task and automatically constructs suitable reward functions and domain randomization distributions to support real-world transfer.
These LLM-derived configurations of reward functions and domain randomization are “competitive with existing human-designed” configurations, so the simulated training process can be automated.
The result is automated training for many robot skills, such as a quadruped balancing and walking atop a yoga ball, all without iterative manual design. The research and the code is open-source.
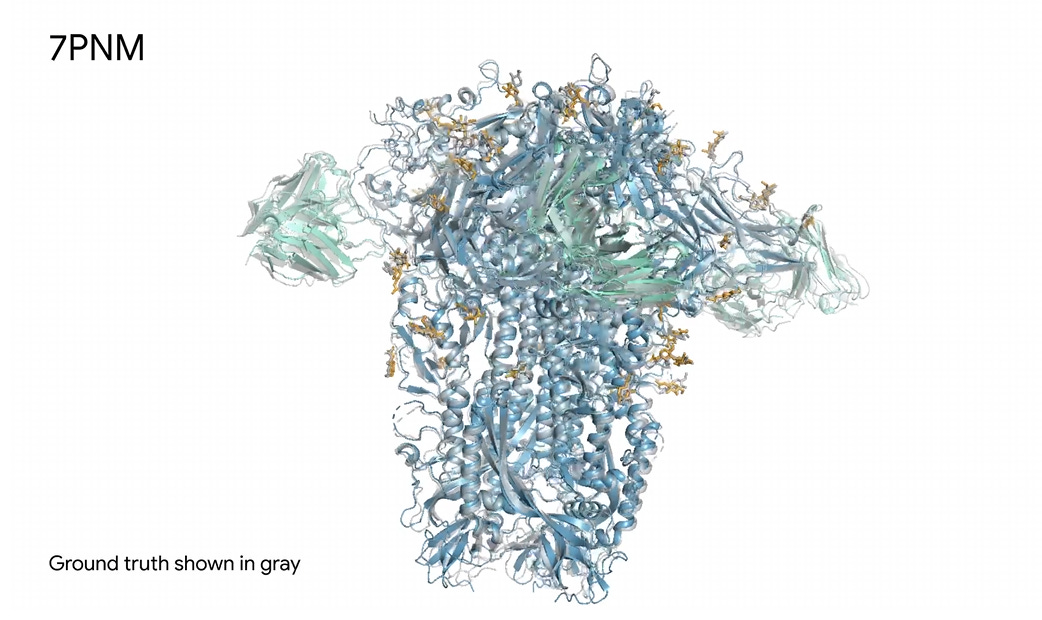
AlphaFold 3: Predicting structure and interactions of all life’s molecules
In a big step forward for biological modeling, Google DeepMind has produced AlphaFold 3, an AI model that expands upon previous work (AlphaFold and AlphaFold 2) to predict the structures and interactions of proteins and other biological molecules:
Given an input list of molecules, AlphaFold 3 generates their joint 3D structure, revealing how they all fit together. It models large biomolecules such as proteins, DNA and RNA, as well as small molecules, also known as ligands — a category encompassing many drugs. Furthermore, AlphaFold 3 can model chemical modifications to these molecules which control the healthy functioning of cells, that when disrupted can lead to disease.
AlphaFold3 is explained further in their blog post “AlphaFold 3 predicts the structure and interactions of all of life’s molecules” and in the paper “Accurate structure prediction of biomolecular interactions with AlphaFold 3” in Nature.
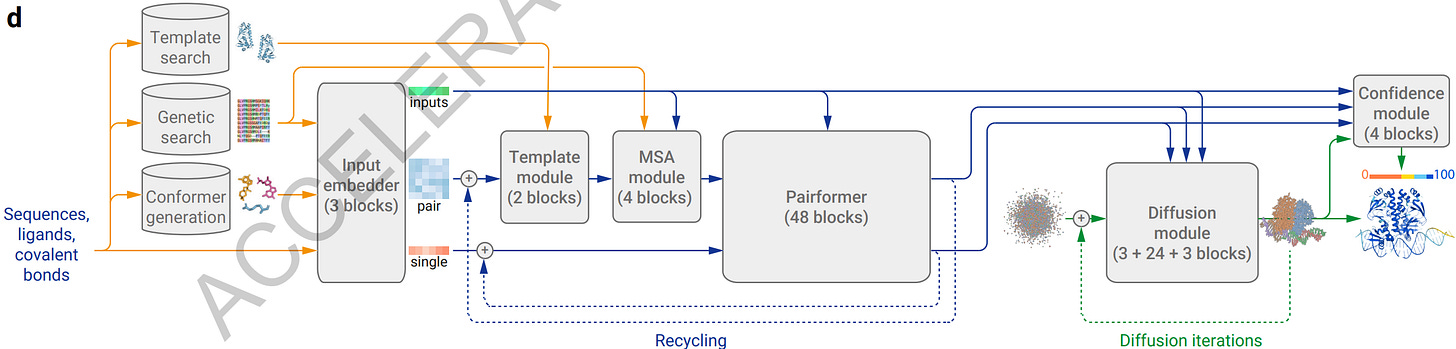
AlphaFold 3 evolves the AlphaFold 2 architecture in various ways, including the addition of a diffusion module, to improve accuracy and accommodate a wider range of chemical entities. As a result:
AlphaFold 3 expands predictions beyond proteins to DNA, RNA, and ligands.
It improves accuracy 50% compared to previous models, “and for some important categories of interaction we have doubled prediction accuracy.”
The original AlphaFold essentially solved a long-standing problem in biology: The protein folding problem. However, real research in medicine, biology, and drug development is concerned not with just single molecule structures, but how different molecules interact.
AlphaFold 3 is an important advance because it brings AI into modelling these interactions between molecules and extends structural understanding across all types of biological molecules.
xLSTM: Extended Long Short-Term Memory
The paper “xLSTM: Extended Long Short-Term Memory” takes an old idea (LSTM, Long Short-term Memory, first) and revives and updates it for the era of LLMs. The result is a modified architecture that beats both Transformer and Mamba architectures in some accuracy metrics.
They notes some weaknesses of original LSTMs as the reason for transformers dominance, specifically in how in handles memory and changes to them, and overcome it by updating LSTMs to create the xLSTM:
Firstly, we introduce exponential gating with appropriate normalization and stabilization techniques. Secondly, we modify the LSTM memory structure, obtaining:
(i) sLSTM with a scalar memory, a scalar update, and new memory mixing,
(ii) mLSTM that is fully parallelizable with a matrix memory and a covariance update rule.
Integrating these LSTM extensions into residual block backbones yields xLSTM blocks that are then residually stacked into xLSTM architectures.
To consider how the new xLSTM architecture scales, they trained xLSTMs, Transformers, State Space Models, and other methods on SlimPajama data (15B and 300B tokens) and compared trained models. They found exponential gating and modified memory structures helped xLSTM perform better than state-of-the-art Transformers and State Space Models, both in performance and scaling.
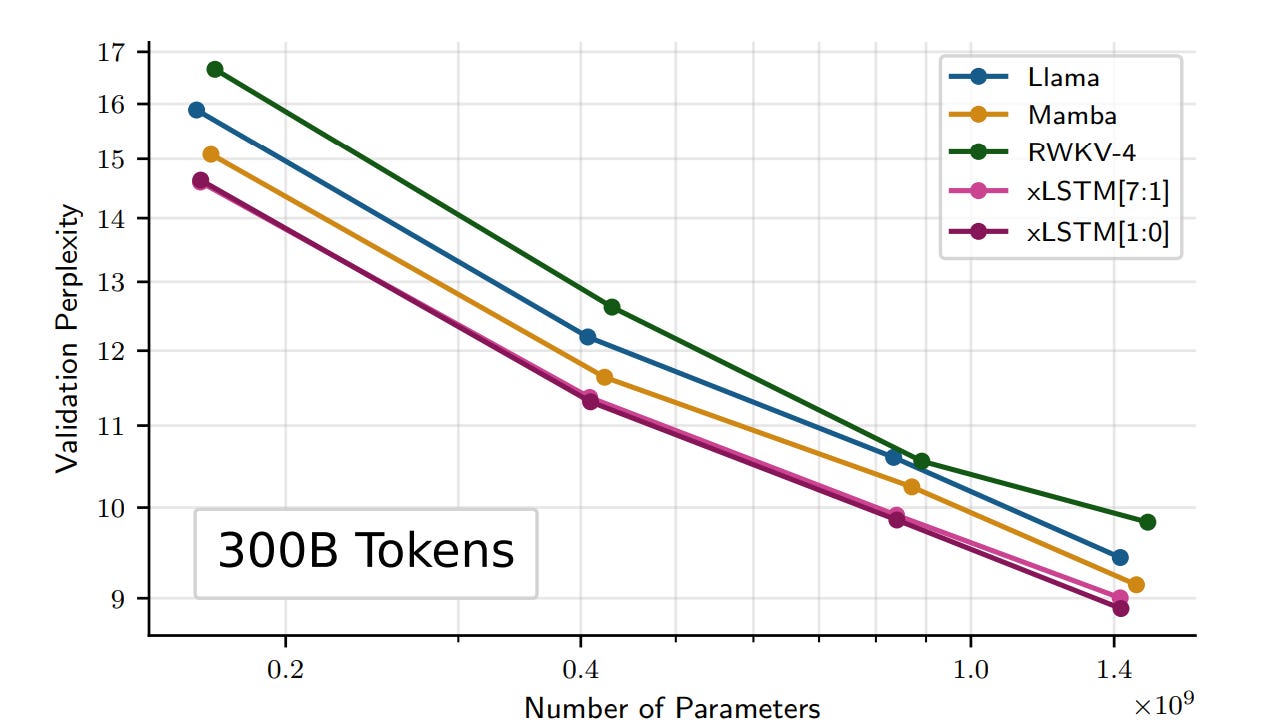
Mamba and similar architectures had a ‘catch’ where more efficient sub-quadratic state-space model suffered an accuracy trade-off versus transformer. Here it has as much flexibility and accuracy in memory as transformers. They do mention limitations and challenges in parallelization, optimization and the computation complexity of memory for mLSTM, and conclude:
We anticipate that an extensive optimization process is needed for xLSTM to reach its full potential.
While xLSTM is apparently efficient enough to be a serious competitor to Transformer architecture, it might not be enough to overcome the inertia Transformers have for LLMs. xLSTM could find a niche in Time Series Prediction or physical systems models, should it prove to be most efficient there.
KAN: Kolmogorov–Arnold Networks
Similar to how xLSTM is presented as an alternative to Transformer, the Kolmogorov–Arnold Networks architecture is presented in “KAN: Kolmogorov-Arnold Networks” as an alternative architecture to MLPs, Multi-Layer Perceptrons.
Multi-layer perceptrons (MLPs) implement fully-connected feedforward neural networks, a vital building block that is pervasive in deep learning. While others have looks at KANs before, this paper generalizes Kolmogorov-Arnold representation to arbitrary widths and depths. Some of the differences:
While MLPs have fixed activation functions on nodes ("neurons"), KANs have learnable activation functions on edges ("weights"). KANs have no linear weights at all -- every weight parameter is replaced by a univariate function parametrized as a spline.
They show that smaller KANs can achieve comparable or better accuracy than much larger MLPs in data fitting and PDE solving, and KANs scale performance faster than MLPs. KANs outperform MLPs in terms of accuracy and interpretability.
One intuition underneath all the math is that “MLPs treat linear transformations and nonlinearities separately as W and σ, while KANs treat them all together in Φ.” They note that “KANs are composed of interpretable functions, so when a human user stares at a KAN, it is like communicating with it using the language of functions.” So KANs are interpretable as functional compositions.
On the devil’s advocate side, there is this: “Kolmogorov-Arnold Network is just an ordinary MLP. … if we consider KAN interaction as a piece-wise linear function, it can be rewritten in MLP terms.” If KANs and MLPs are different implementations of functional decompositions, deciding which to use is a matter of practicality. However, KANs have a big practical downside, as the authors admit:
Currently, the biggest bottleneck of KANs lies in its slow training. KANs are usually 10x slower than MLPs, given the same number of parameters.
While this KAN result is an interesting and potentially important theoretical result, it won’t displace MLPs due to that 10x gap; making KANs a practical alternative to MLPs remains future work.
VoT: Visualization of Thought
The paper “Visualization-of-Thought Elicits Spatial Reasoning in Large Language Models” proposes the prompting technique Visualization-of-Thought (VoT) to enhance LLMs' spatial reasoning, enabling them to outperform MLLMs in spatial tasks.
As with Chain-of-Thought, Visualization-of-thought (VoT) breaks down a reasoning process into steps, in this manner:
• GPT-4 CoT: Let’s think step by step.
• GPT-4 VoT: Visualize the state after each reasoning step.
VoT’s prompting approach elicits the "mind's eye" of LLMs by specifically reasoning on spatial tasks. When tested on multi-hop spatial reasoning tasks like visual tiling and visual navigation, VoT outperforms existing multimodal LLMs.
Visualization-of-Thought has utility for LLMs interacting with or understanding graphical interfaces. PyWinAssistant, a Large Action Model that controls human user interfaces via natural language, was recently released. It utilizes VoT to perform its tasks.
The success of this suggests there may be more cognitive abilities yet to explore based on “X of Thought” techniques, to elicit more LLM and multimodal LLM capabilities.
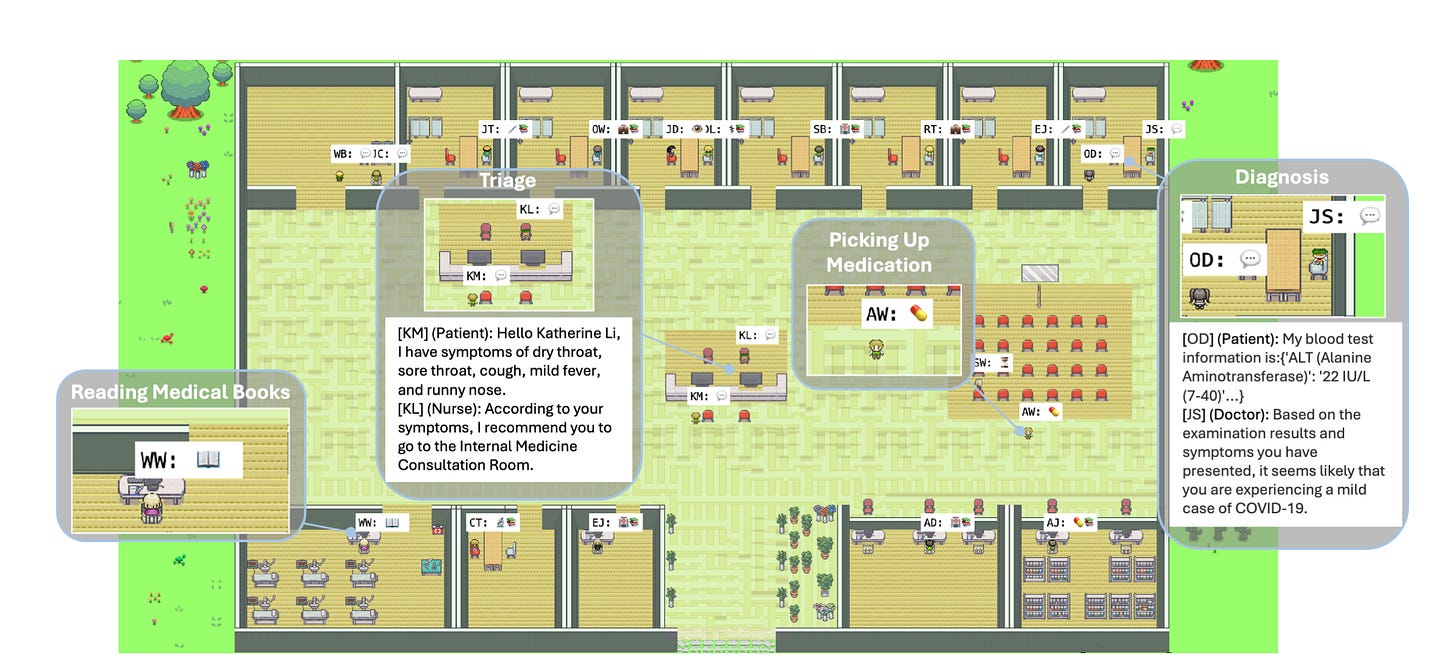
Agent Hospital: Simulating a Hospital with Medical Agents
You may recall there was a paper last year that simulated a town with AI Agents. The paper “Agent Hospital: A Simulacrum of Hospital with Evolvable Medical Agents” has taken that idea into a hospital environment.
In this environment, the patients, nurses, and doctors are autonomous agents powered by LLMs, with the goal of using this environment to train AI agents to diagnose illness:
As the simulacrum can simulate disease onset and progression based on knowledge bases and LLMs, doctor agents can keep accumulating experience from both successful and unsuccessful cases.
Doctor agents learn to make better decisions through simulated interactions, and also “the knowledge the doctor agents have acquired in Agent Hospital is applicable to real-world Medicare benchmarks.” Specifically:
the evolved doctor agent achieves a state-of-the-art accuracy of 93.06% on a subset of the MedQA dataset that covers major respiratory diseases.
This shows applicability to training virtual AI agents in simulated environments.